Linear Absorption Coefficient (μ) Examples
These examples will demonstrate how to calculate the Linear absorption
coefficient, μ, using different methods provided in diffpy.utils.
Methods for obtaining linear absorption coefficient
Obtaining μ can be done in two
different ways using diffpy.utils.
Using a “z-scan” measurement: Perform a z-scan measurement on the sample and use
diffpy.utils.tools.compute_mudto calculate μ.Using tabulated values: Given composition, density, and X-ray energy, use
diffpy.utils.tools.compute_mu_using_xraydbto calculate μ from tabulated values.
Why is μ Important?
The linear absorption coefficient, μ, quantifies how much X-ray radiation is absorbed by a material per unit length. It is a critical parameter in many scientific techniques.
For example, when calculating pair distribution functions (PDFs)
using diffpy.pdfgetx,
a key assumption is that the linear absorption is negligible.
This is frequently the case for high-energy X-rays. However,
this must be corrected for when using low energy X-rays, such
as those from a laboratory source. To correct for linear absorption,
the linear absorption coefficient, μ, must be known.
Correcting for linear absorption with diffpy.labpdfproc
If your objective is to correct for linear absorption in PDF calculations,
please refer to our package diffpy.labpdfproc. This package is specifically
designed to correct your laboratory X-ray PDF data for absorption effects.
More information can be found in the
diffpy.labpdfproc documentation.
Calculating μ from a “z-scan” Measurement
Note
The data we will be using for this example can be found, here.
A “z-scan” measurement is the measured transmission of your X-ray incident beam as a function of sample position. This is obtained by moving the sample perpendicular to the X-ray beam (z-direction) and recording the transmitted intensity at each position. This measured data looks something like this,
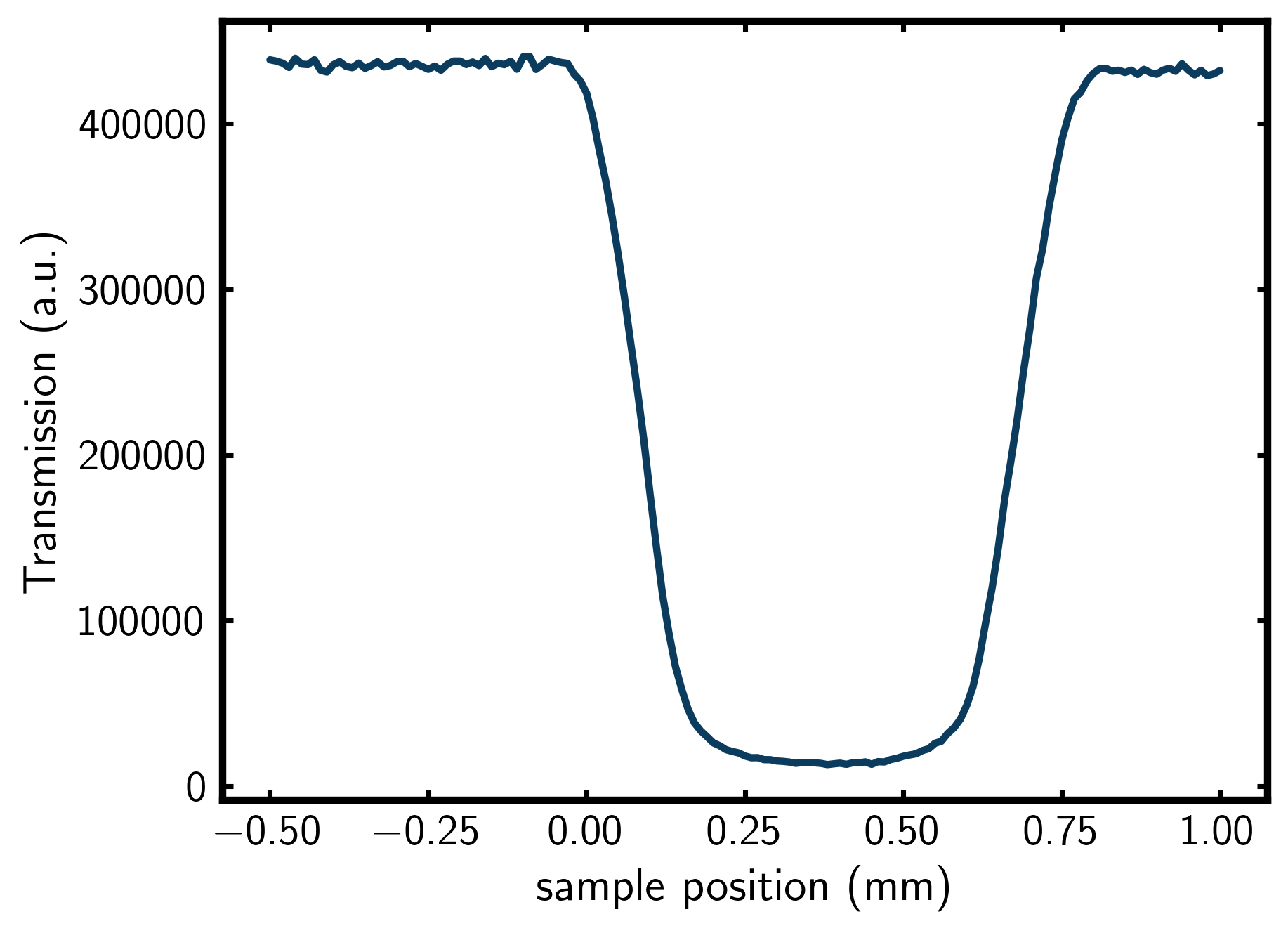
Using this z-scan data, you can calculate μ·d, where d is the inner diameter of
your sample capillary. To do this, simply pass your z-scan measurement to the compute_mud
function from the diffpy.utils.tools module.
First, import the compute_mud function,
from diffpy.utils.tools import compute_mud
Next, pass the filepath to the function,
filepath = "CeO2_635um_zscan_200umSlit_chanClose_exported.xy"
capillary_diameter = 0.5 # mm
mud = compute_mud(filepath)
print(f"Calculated mu*d: {round(mud, 3)}")
print(f"Calculated mu: {round(mud / capillary_diameter, 3)} mm^-1")
This will output the calculated value of μ·d, which is unitless, and μ in mm-1.
Calculated mu*d: 3.489
Calculated mu: 6.977 mm^-1
Calculating μ from Tabulated Values
The function to calculate μ from tabulated values is located
in the diffpy.utils.tools module. So first, import the function,
from diffpy.utils.tools import compute_mu_using_xraydb
To calculate μ, you need to know the sample composition, and X-ray energy, and sample mass density (g/cm3).
composition = "Fe2O3"
energy_keV = 17.45 # Mo K-alpha energy
sample_mass_density = 5.24 # g/cm^3
Now calculate μ using the compute_mu_using_xraydb function.
mu = compute_mu_using_xraydb(composition, energy_keV, sample_mass_density)
print(f"Calculated mu: {round(mu, 3)} mm^-1")
This will output the calculated linear absorption coefficient, μ, in mm-1.
Calculated mu: 13.967 mm^-1